
Either the matrix is not PSD, or there was an issue while computing the eigendecomposition of the matrix.Įrror message (2): ValueError: Precomputed metric requires shape (n_queries, n_indexed). PrincipalDf = pd.DataFrame(data = PrincipalComponents, columns = )įinalDf = pd.concat(]], axis = 1)Įrror message (1): ValueError: There are significant negative eigenvalues (1.11715 of the maximum positive). Principal component analysis (PCA) is a classic multivariate technique that analyses a data table where observations are described by several inter-correlated quantitative dependent variables.16,17PCA of the data is performed to understand the factorsaectingthespectralvariationacrossthesamples,andit is calculated from the covariance matrix of t. PrincipalComponents = pca.fit_transform(X_std)
#Pca method for hyperimage code
Please find the code and error message below.Ĭode: from composition import PCA, KernelPCA, SparsePCA, IncrementalPCAįrom hyperopt import hp, tpe, atpe, fmin, Trials, rand, STATUS_OK I know that KPCA does not have a score in order to find the accuracy of the PCA model, so, how can I overcome this error? I tried several scoring methods and either I get an error from inverse_fit or the size of the array. I tried to code and combine the hyperopt code with KPCA, but, I keep on getting errors at the area dealing with scoring of the PCA model. Principal component analysis (PCA) is widely used for data reduction in group independent component analysis (ICA) of fMRI data. Nonnegativity-constrained multivariate curve resolution via the.
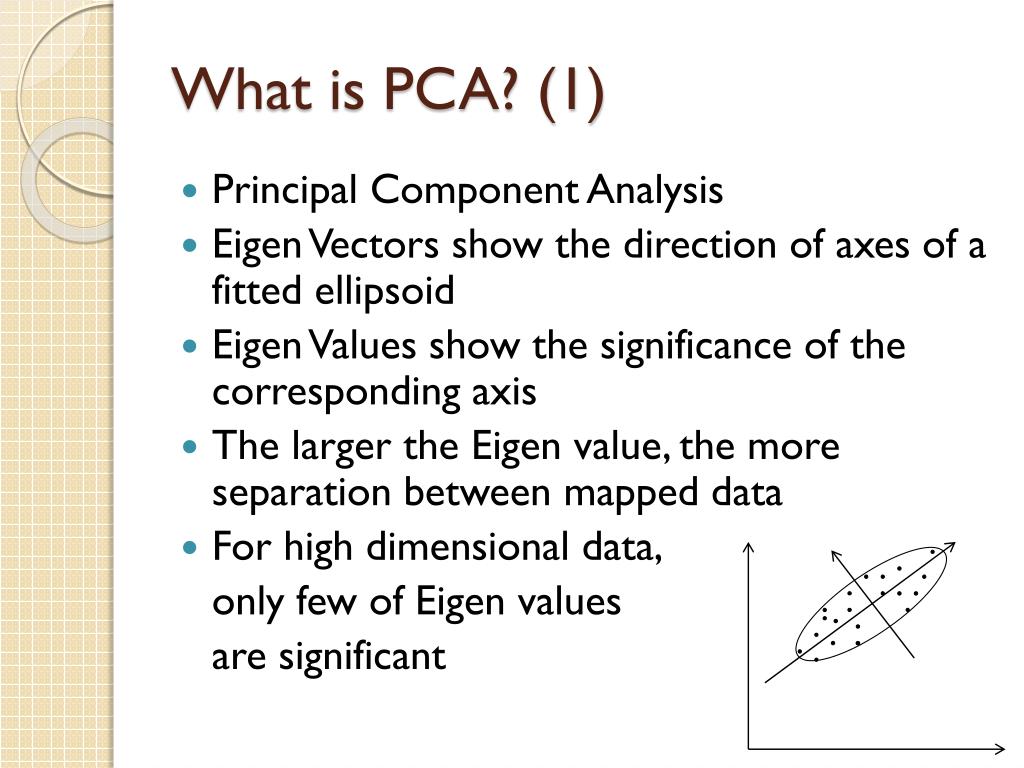
HyperOpt: Bayesian Hyperparameter Optimization Principal component analysis (PCA) is used as both a data reduction and de-noising method in group independent component analysis (ICA) (Calhoun et al.
Furthermore, I went through the following links that look into the hyperparameters method used for classification models: I went through the parameters used in KPCA in scikit learn package and understood that there are some parameters that should work if one of them is selected (For instance, if gamma is selected then degree and coefficient are not used). I am looking into applying Kernel Principal Component Analysis (KPCA) to reduce the dimensionality of my feature matrix set to obtain a cluster of datapoints.


 0 kommentar(er)
0 kommentar(er)
